ML with Python on HPC
Why use Python?
- Python is probably the most popular language for machine learning
- All popular machine learning platforms provide Python APIs
- All HPC consultants are very familiar with Python
(minor) Caveats
- There are a lot of moving pieces:
- Multiple Python versions
- Multiple machine learning platforms
- GPUs
- Jupyter
- Conda 😭
Setting up Python for ML on HPC
- It is best to use Python or Conda virtual environments (venvs) to install your packages
- To create venvs with Python:
module load python/3.8 module load cuda11 cuda11-dnn cuda11-sdk python3 -m venv --system-site-packages <path-to-venv> source <path-to-venv>/bin/activate pip3 install <packages separated by space> pip3 install jupyter --force-reinstall ipython kernel install --name <kernel-name> --user \ --display-name <optional-display-name> - To create venvs with Conda, check HPC docs
Access Jupyter from OOD
Navigate to https://ood.hpc.arizona.edu/. After login, you will see the Open OnDemand dashboard.
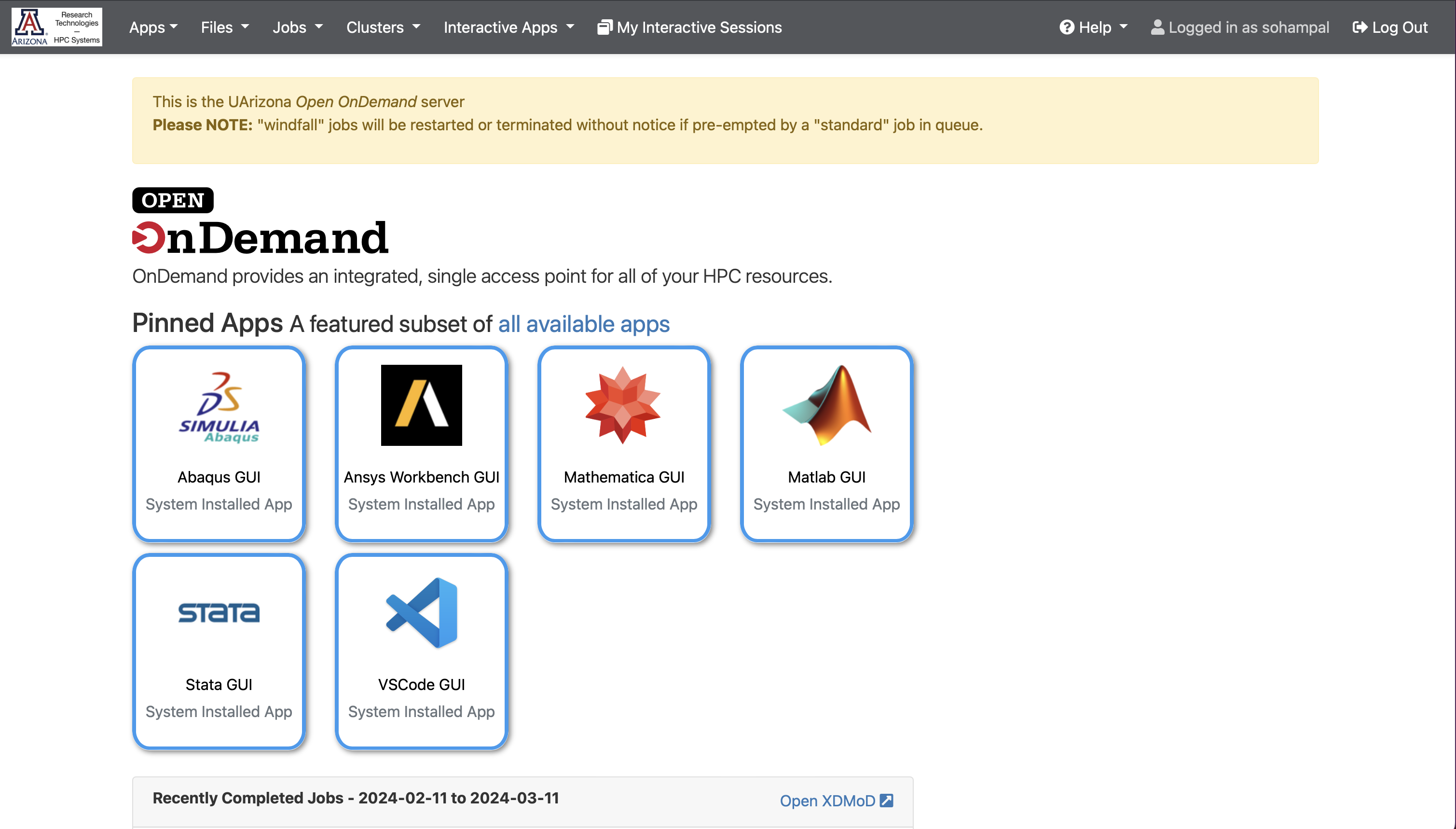
Select Interactive Apps, and then from the drop-down menu select Jupyter Notebook.
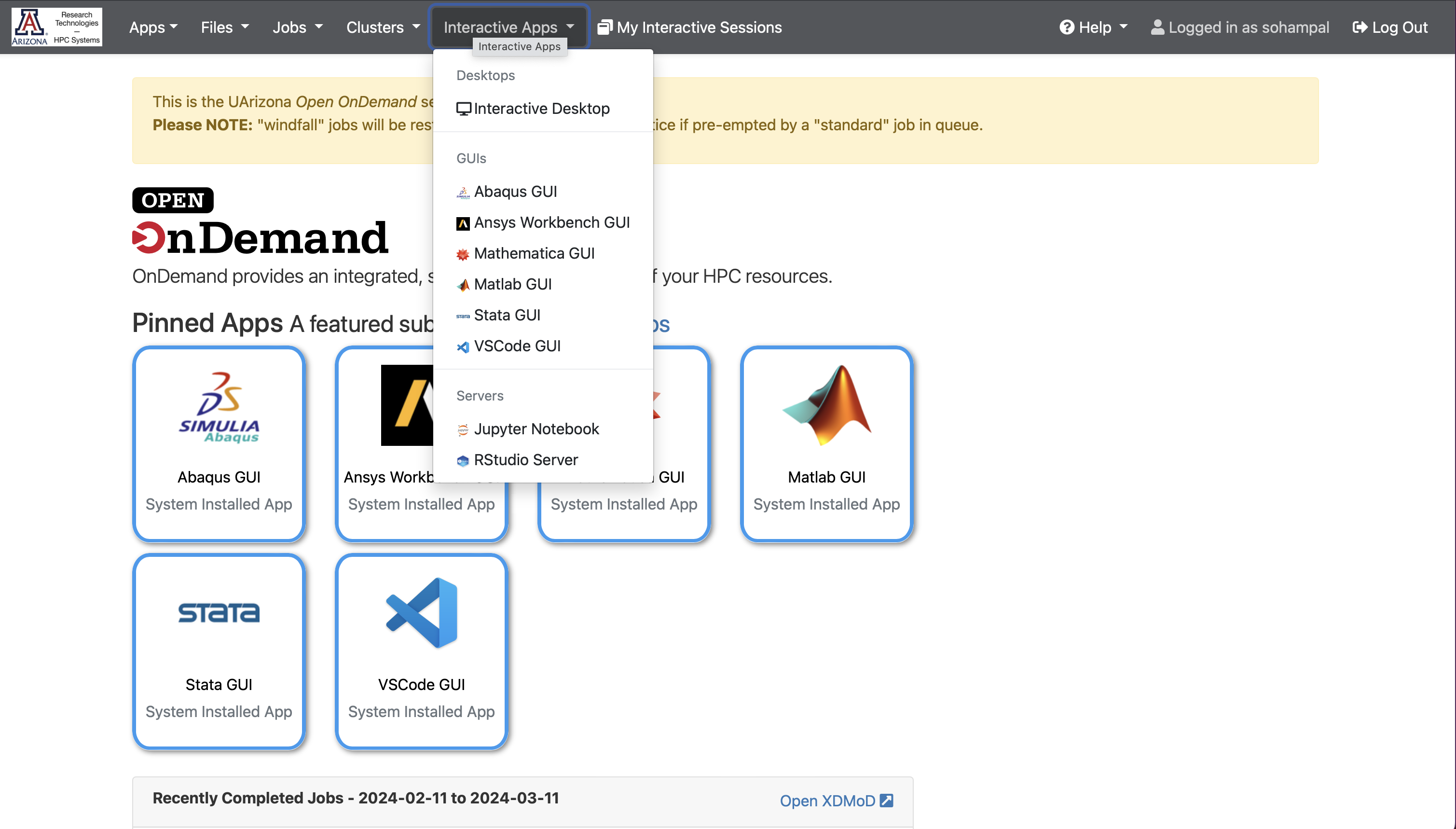
Fill in the details in the form that opens up, and select Launch.

After the session becomes available, select Connect to Jupyter.
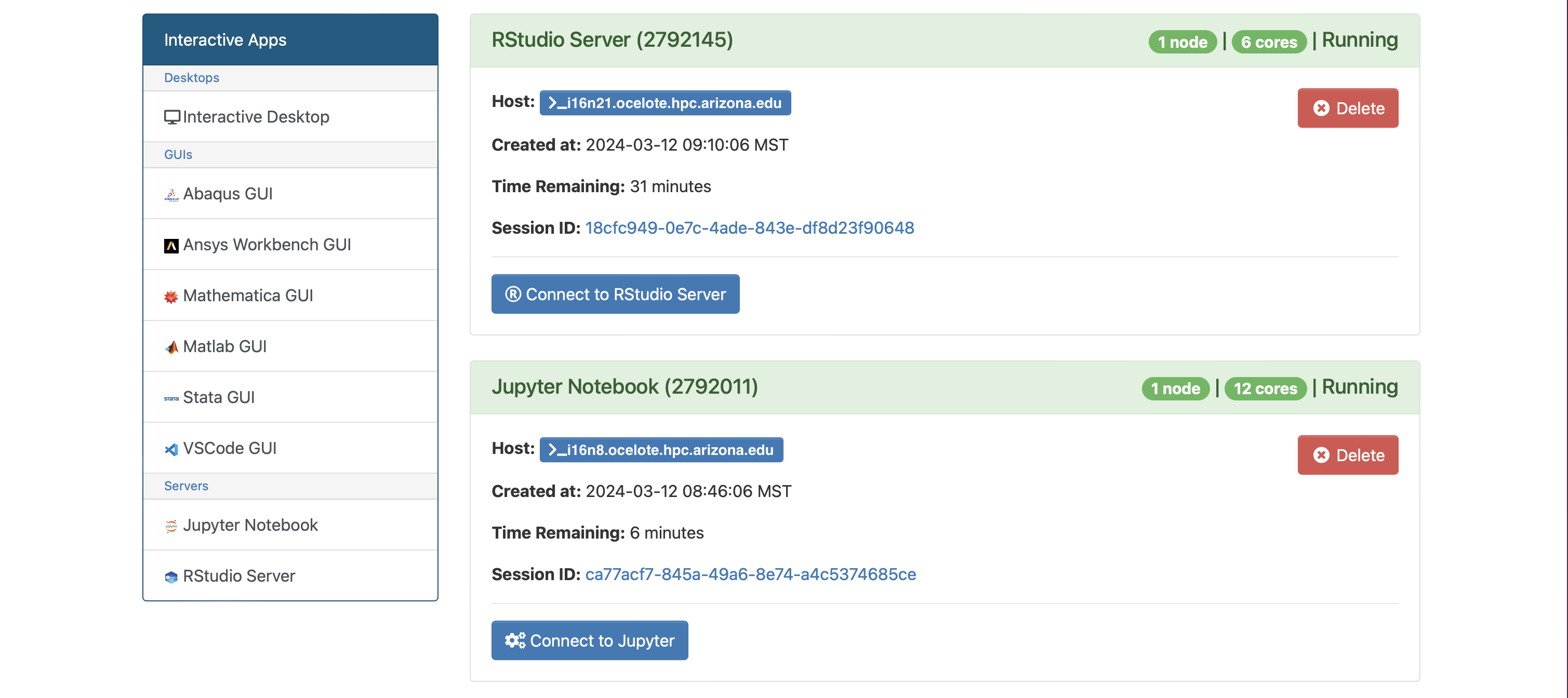
Examples
For today’s examples, install torch, torchvision, torchaudio and fastai.
- Pip 😊
pip3 install torch torchvision torchaudio \ --index-url https://download.pytorch.org/whl/cu118 pip3 install fastai - Conda 🙁
conda install pytorch torchvision torchaudio \ pytorch-cuda=11.8 -c pytorch -c nvidia conda install -c fastai fastai
🍄 classification
Identify fungi species by their images

Full dataset: Danish Fungi 2020
- Local dataset: Four species with the most images in the DF20-Mini dataset
- You can copy the data to your working directory (under your home directory, or your PI’s
/groupsor/xdiskshare), and untar it:cd <working-dir> cp /contrib/datasets/workshops/DF20M-4.tar.gz ./ tar xvf DF20M-4.tar.gz
Download the Jupyter notebook: fungi-classification.ipynb
Clustering 🐧
Cluster penguins into groups based on their bill features

Artwork by @allison_horst
- We will use mean shift clustering to cluster the penguins
- Mean shift clustering updates candidates for centroids to be the mean of the points within a given region
- Each observation belongs to the cluster with the nearest mean (centroid)
- You have to provide the bandwidth of the kernel
- Penguin data comes from the
palmerpenguinsdataset
Download the Jupyter notebook: penguins-classification.ipynb